Molecular similarity network with visualised structures
Published:
Analogous to a Sequence Similarity Network (SSN), molecular similarity network visualises Tanimoto similarity between molecules. iwatobi shows how to construct one with molecular structures here and here using Python packages RDKit, networkx, and cyjupyter.
Here is my own Jupyter notebook with a little bit of refactoring and more comments to improve the readability (I rarely use RDKit so this is more for my benefit, really). Instead of SMILES in the graph, I use the compound names instead. Here’s how 20 compounds would look like:
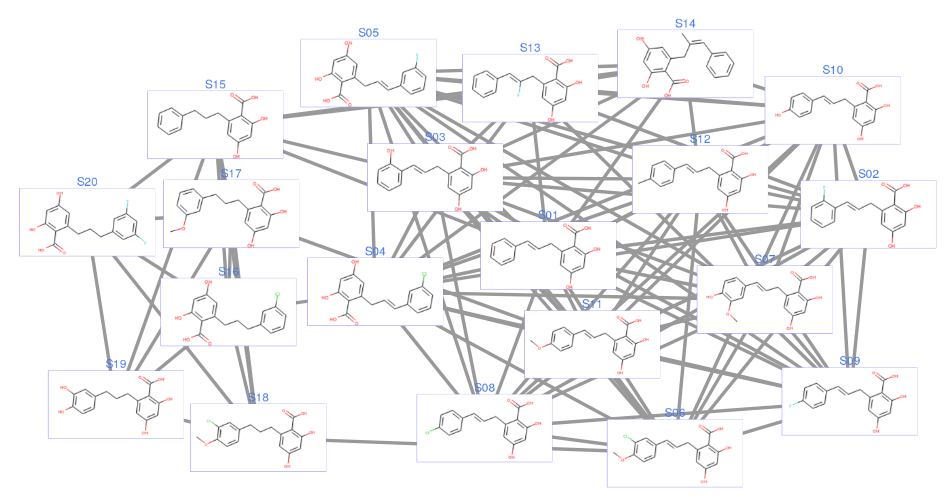
The only problem I see with this is scalability. For around 20 molecules or so this approach is good. More than that, the graph is too crowded with molecular structures and the clustering would not look obvious.

Leave a Comment
Your email address will not be published. Required fields are marked *